library(tidyverse)
library(lmerTest)
library(broom)
library(broom.mixed)マルチレベルモデルにおけるよくある誤解
「マルチレベルモデルでやんなきゃ係数にバイアスが…」←ホント?
データ生成
マルチレベルのデータを考える サンプルサイズ1000、グループ数50のデータ
\[\begin{align} y_{ig} \sim 0.5x_{ig} + \mathrm{Normal}(\theta_g, 1) \\ \theta_g \sim \mathrm{Normal}(0, 3) \\ x_i \sim \mathrm{Normal}(0, 1) \end{align}\]
dgp <- function(samplesize = 1000) {
tibble(
id = 1:samplesize,
group = rep(1:50, 20),
x = rnorm(samplesize, mean = 0, sd = 1),
) |>
group_by(group) |>
mutate(group_mean = rnorm(1, mean = 0, sd = 3)) |>
ungroup() |>
mutate(y = 0.5*x + rnorm(samplesize, mean = group_mean, sd = 1))
}
data <- dgp()lm(y ~ x, data = data) |>
summary()
Call:
lm(formula = y ~ x, data = data)
Residuals:
Min 1Q Median 3Q Max
-9.9673 -2.3079 0.2522 2.4065 7.8351
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.01677 0.10746 0.156 0.8761
x 0.21591 0.10766 2.005 0.0452 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 3.398 on 998 degrees of freedom
Multiple R-squared: 0.004014, Adjusted R-squared: 0.003016
F-statistic: 4.022 on 1 and 998 DF, p-value: 0.04518lmer(y ~ x + (1|group), data = data) |>
summary()Linear mixed model fit by REML. t-tests use Satterthwaite's method [
lmerModLmerTest]
Formula: y ~ x + (1 | group)
Data: data
REML criterion at convergence: 3144.8
Scaled residuals:
Min 1Q Median 3Q Max
-3.4810 -0.6754 0.0040 0.7066 3.4708
Random effects:
Groups Name Variance Std.Dev.
group (Intercept) 10.767 3.281
Residual 1.038 1.019
Number of obs: 1000, groups: group, 50
Fixed effects:
Estimate Std. Error df t value Pr(>|t|)
(Intercept) 0.01684 0.46517 48.99703 0.036 0.971
x 0.46739 0.03312 949.48170 14.111 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Correlation of Fixed Effects:
(Intr)
x 0.000 シミュレーション
データを1000個作成
data_list <-
map(1:1000, \(x) dgp(1000)) |>
enframe()OLSと変量効果モデルで推定
result <-
data_list |>
mutate(
lm = map(value, \(data) lm(y ~ x, data = data)),
lmer = map(value, \(data) lmer(y ~ x + (1|group), data = data))
)xの係数のみ取り出す
res2 <-
result |>
mutate(
lm_res = map(lm, \(model) {
tidy(model) |>
select(term, estimate)
}),
lmer_res = map(lmer, \(model) {
tidy(model) |>
select(term, estimate)
})
) |>
select(name, lm_res, lmer_res) |>
pivot_longer(!name, names_to = 'model', values_to = 'value') |>
unnest(value) |>
filter(term == 'x')結果を図示
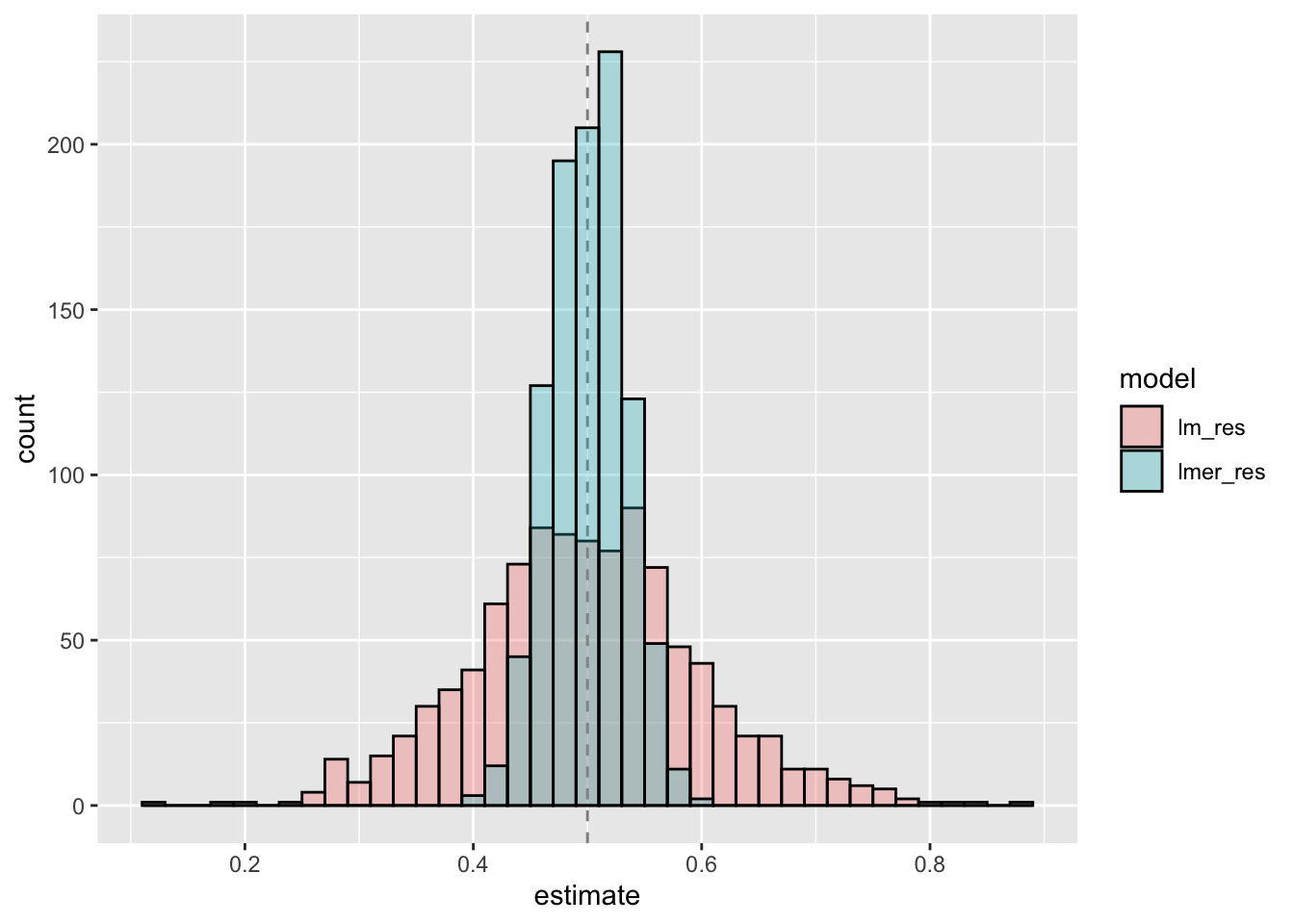
- どちらの点推定値も真の値の0.5を中心に分布=バイアスはない
- OLSによる点推定値はバリアンスが大きい
- 変量効果(マルチレベル)モデルによる点推定値はバリアンスが小さい
res2 |>
ggplot(aes(estimate, fill = model))+
geom_vline(xintercept = 0.5, linetype = 'dashed', alpha = 0.5)+
geom_histogram(alpha = 0.3, color = 'black', binwidth = 0.02, position = 'identity')+
scale_x_continuous(breaks = seq(0, 1, 0.2))